Introduction
A few weeks ago, the New York Times published an article titled A.I. Is Learning What It Means to Be Alive. I'm not the biggest fan of the title, but I am happy the subject in it is being talked about more! It laid out the story of how so-called 'scRNA-seq Foundation Models' may potentially change how single-cell RNA sequencing (scRNA) data is interpreted, used, and applied. Though the Times article was fantastic in its own right, it asks very surface-level questions about the whole process. I'd like to do a much deeper dive in this topic and try to walk through the motivation, ideas, and process of creating models, along with what they do well on and what they still struggle with.
Warning in advance: single-cell biology is complex to the extreme and pretty much every facet of it is hotly debated. This means this post is partially an interpretation, meant to offer some wider background context on the field, and may strongly deviate from others perspectives when it comes to specifics.
scRNA-seq
What is scRNA-seq?
Let's come up with a goal for ourselves: we'd like to learn the cellular identity of every cell in the human body. Why? Well, for knowledge sakes to start off with, we'll come up with a concrete use-case later.
First question: what exactly is cellular identity?
Maybe cell types? Let's consider an organ, like the brain. You may recall there are different cell types in the brain, neurons, glial cells, oligodendrocytes, and so on. But what are the functional differences between these cells, upon what means have scientists chosen their 'type'? One way that we often rely on is the proteins they have; the proteins that live within the cells, the proteins that they put on their surface, the proteins that they secrete. Cells that share many of the same proteins could serve as a notion of cellular identity. Indeed, proteins are involved in an awful lot of processes: how cells communicate with cells around them, how cells intake in resources, how cells respond to environmental stimuli, and many more. One could make the argument that proteins aren't actually the true differentiator, potentially cell metabolites or epigenetic modifications are more important. Well, we could discuss that in a future post. But let's move on with the axiom 'cell state could be best understood as the proteins they have'.
An example is syncytiotrophoblasts, which are cells found in placental tissue and are involved in nutrient transfer from maternal blood to fetal capillaries. As such, you may naturally expect that they express proteins specialized in nutrient transfer. And, indeed, they express TFRC (Transferrin receptor) at an extremely high rate compared to almost all other cells in the human body, TFRC is mainly involved in intaking in iron, and iron is an extremely important building block for crafting a living thing from scratch.
I've given quite a neat example, but is it always this way? Unfortunately, not every protein has such a clear-cut role, many of them are so-called 'housekeeping proteins' (or, more specifically, housekeeping genes) that are essential for the ongoing maintenance of any cell, no matter what they do. The canonical example are proteins involved in cell division, such as Cyclin-H, without which your cells would be entirely incapable of reproducing. But still, a significant fraction of all proteins in the human body do have extremely unique functionality well-suited to the cells that produce them, enough to where we could potentially arrive to a notion of cell identity purely from observing where certain proteins do and do not appear.
And, with that, we run into our first challenge: how exactly do we assess protein content of any given cell? There's a few things we could try. Feel free to skip this section if it gets a bit much! Spoiler alert: none of them will work for us anyway.
Immunocytochemistry. The basic idea is that we create a special protein (an antibody) that's designed to stick to the protein we're interested in, potentially the TFRC from before. We also attach a little chemical tag (fluorophore) to the antibody that lights up when a specific wavelength of light is shined on it. Now, we simply take our cells and run the antibody over them. Using the TFRC example, the antibodies will diffuse through the cell, hunting for TFRC proteins. When an antibody finds a TFRC molecule, it'll tightly bind to it. The final step is to look at the cells under a fluorescence microscope. This type of microscope shines light of a specific wavelength onto the cells, exciting the fluorophores. The fluorophores emit light in response, revealing the location of the TFRC proteins. If TFRC is primarily located on the cell surface, for example, we would see a bright outline around each cell.
Flow Cytometry: This technique is similar to immunocytochemistry in that it uses antibodies to detect specific proteins. However, instead of looking at the cells under a microscope, flow cytometry uses a specialized instrument called a flow cytometer to analyze the cells one by one. We start by creating a single-cell suspension, meaning that the cells are no longer stuck together in a tissue but are floating freely in a liquid. We then incubate these cells with antibodies that are specific to the protein we're interested in (like TFRC). These antibodies, as before, are typically connected to a fluorophore and will bind to our suspended cells. The cell suspension is then run through the flow cytometer. Inside this machine, the cells are forced to pass through a narrow channel one at a time. As each cell passes through, a laser beam is shone onto it. If the cell has been labeled with a fluorescent antibody, the laser will cause the dye to emit light. The flow cytometer detects this light and can quantify how much of the protein is present on each cell.
Mass spectrometry: Here, we essentially take a cell, break it open, and then analyze all the proteins within it to get a picture of what proteins were there to begin with. We first chop the cellular proteins up into smaller pieces using enzymes. For example, we may use 'trypsin', which cuts proteins whenever it encounters the amino acids lysine or arginine. The resulting pieces, which we call peptides, are typically around 10-20 amino acids long. We'll then load the mixture of peptides into a liquid chromatography (LC) system, which separates the peptides based on their chemical properties. As the peptides exit the LC system, they are sprayed into the mass spectrometer using a process called electrospray ionization (ESI). This process converts the peptides into charged particles (ions).Once inside the mass spectrometer, the peptide ions are further separated based on their mass-to-charge ratio (m/z) in a component called the mass analyzer. There are different types of mass analyzers, but they all use electric and/or magnetic fields to manipulate the path of the ions, Because all the ions are given the same kinetic energy by the electric field, their velocity (and thus the time it takes them to reach the detector) depends on their mass. Lighter ions will reach the detector faster than heavier ones. As the ions reach the detector, their m/z and abundance are recorded. This data is then processed to identify which peptides were present and, by extension, which proteins those peptides originated from. By comparing the identified peptides to databases of known protein sequences, we can determine which proteins were present in the original sample and even quantify their relative abundance.
I've already given it away; despite the power of these methods, none of will adequately work for us in our lofty goal to learn about the protein landscape of every cell in the human body.
Immunocytochemistry is a time-consuming process, requires known antibodies that bind to known proteins (which not every protein has!), and extremely low throughput (so measuring even a thousand cells is an undertaking). Flow cytometry, while higher throughput, is similarly dependent on known antibodies, and does not work for tissues that cannot be easily suspended, such as brain tissue or adipose tissue (both due to their inherently fragility). Finally, mass spectrometry, while not relying on known antibodies, requires an enormous number of cells to capture an accurate readout of protein content (which means we lose out on understanding single cells), may miss out of rarer proteins, and is by far the most time-consuming method in this list. Of course, I'm oversimplifying, all of these methods likely have some alternative protocol which somewhat solves these problems, but even those remain insufficient.
So, now what? Unfortunately, there isn't really a backup, learning proteomic landscapes of cells in a way that is scalable to the millions, allows us to discover rare proteins, and has a single-cell-resolution is, thus far, an unsolved problem. Let's think backwards; if we cannot have a perfect view of proteins in a cell, perhaps we can settle for measuring the things that cause the proteins to exist at all?
Of course, as the title of this post may suggest, we're talking about RNA, specifically messenger RNA (mRNA). mRNA is created (more specifically, 'transcribed') directly from a cells DNA and is transported to the ribosome, where each snippet of mRNA is interpreted (3 nucleic bases as a time) to create a single amino acid, which is chained together in order. For example, if an mRNA segment is composed of UUU-GUA-CCA, that is mapped to a protein of amino acids Phe-Val-Pro.
If we measure the sum amount of all mRNA that is in this intermediary zone between transcribed and translated at a given point in time, or its 'transcriptional landscape', this indirectly also gives us the proteomic landscape, correct? Well, not exactly.
Not all mRNAs are translated into proteins at the same rate or efficiency. Some mRNAs are highly stable and can persist in the cell for hours or even days, while others are rapidly degraded. Which means that mRNAs are translated multiple times by ribosomes, yielding many copies of the corresponding protein, others may only be translated once or not at all. This means that the relative abundance of mRNAs in a cell doesn't always directly correspond to the relative abundance of their encoded proteins. Similarly, protein half lives dramatically vary; proteins can persist for days or even weeks. This means that the protein composition of a cell can reflect its past transcriptional states, not just its current one. A protein that was abundant in a cell yesterday could still be present today, even if its associated mRNA has been degraded. Conversely, a newly transcribed mRNA may not yet have been translated into a detectable amount of protein. Finally, mRNA do not map to proteins perfectly, plenty of proteins undergo 'post-translational modification', which means that the protein you end up with from a given mRNA in a given environment may have a completely different structure than the same mRNA in a different environment.
Yet, for all the nuanced problems that the relationship of mRNA and protein expression has, they do have a connection. A 2016 review paper from Cell titled 'On the Dependency of Cellular Protein Levels on mRNA Abundance' has this to say about it:
At Steady State, mRNA Levels Primarily Explain Protein Levels
It is challenging to rigorously define the term ‘‘steady state’’ for cells, especially if they are undergoing long-term dynamic processes such as continuous proliferation (Hsieh et al., 2012), differentiation (Kristensen et al., 2013), or other types of fate decisions (Lu et al., 2009; Gru¨ n et al., 2014). However, large ensembles of cells as they are typically used for ‘‘omics’’ experiments can be regarded being at steady state if the average protein and/or mRNA levels remain relatively stable over time (normally above several hours). Numerous studies published during the last 15 years suggest that, under such circumstances, gene-to-gene variation of protein levels is primarily determined by their respective mRNA levels.
A fair bit of the evidence comes from a relatively well-known 2011 paper from Schwanhäusser et al. that studies fibroblast mouse cells, concluding that the variance of mRNA explains about 40% of protein expression levels. From the same review paper:
[Schwanhäusser] investigated the relationship between mRNA and protein levels in unperturbed mammalian cells using RNA-seq, pSILAC, and absolute quantification strategies. His study determined that about 40% of the variance of protein levels between different proteins could be explained by mRNA levels (coefficient of determination, i.e., squared Pearson correlation coefficient between mRNA and protein abundances, R2 = 0.41). A follow-up study re-analyzing the same dataset with a different statistical model concluded that about 56%–84% of the protein variance could be explained by mRNA variance...
Another complicating factor is that one study finds that it's very tissue dependent, with the explained variance range being 46-68%. But, overall, we're in a good spot. Given that we cannot easily access protein expression en-masse, gathering together mRNA transcripts is seeming like it could be a decent approximation of cell identity. But...can we do that?
We can! And finally, we can start talking about the title of this subsection: scRNA-seq, or single-cell transcriptomic sequencing. This method solves basically every problem that the protein expression ones had: it is massively scalable, capable of capturing single-cell-level resolution, and can find rare transcripts (and thus proteins).
Finally, we'll end this section with a simple question: how does it work? scRNA-seq is, in my opinion, the most conceptually complicated of the methods we've discussed so far. Again, the explanation of this method isn't actually important, but is included here for completeness, it can be skipped. Here's the general workflow, and also an excellent youtube video that goes over the same stuff:
Cell Isolation: The first step is to obtain a single-cell suspension from the tissue or sample of interest. This often involves enzymatic digestion or mechanical dissociation to break down the extracellular matrix and separate the cells from each other. The goal is to have a suspension where each cell is floating freely.
Cell Capture: Next, individual cells need to be isolated into separate reaction chambers for further processing. There are several methods for this, including microfluidic devices, microdroplet-based methods, or microwells. Each of these methods aims to seperate single cells from one-another.
Cell Lysis and RNA Capture: Once individual cells are isolated, they are lysed (their cell membranes are disrupted) to release their RNA content. The RNA is then captured, typically using oligo-dT primers that bind to the poly-A tails of mRNA molecules. In other words, the end-piece of every cell's RNA is labeled with a unique molecular identifier (UMI) and a cell-specific barcode. The UMI allows for the correction of amplification bias during the next step, while the cell barcode allows the RNA to be traced back to its cell of origin. This allows us to have single-cell level resolution!
Reverse Transcription and Amplification: The captured RNA is then reverse transcribed into cDNA, which is more stable and can be amplified. The cDNA is amplified using PCR, generating many copies of each cDNA molecule. This is what allows rare transcripts to be detected! We're no longer limited by the base level of mRNA in a cell, it can be scaled up!
Library Preparation and Sequencing: The amplified cDNA is then sequenced as normal by Next-Generation-Sequencing (NGS) platforms that work with DNA. This is what allows massive scalability, NGS platforms are able to operate on a scale of millions of genetic fragments of once. There is a step afterword for correcting errors in this process, such as removing mitochondrial DNA that we typically don't care about, but we can skip that as it's not super relevant here.
And what does the end result of scRNA-seq look like? Simple, and one that's familiar to anybody who has worked with dataframes before: a 2 dimensional count matrix, where each column represents a cell, each rowrepresents a gene, and each entry represents the expression level of a particular gene in a particular cell in terms of 'number of UMIs', or transcripts, found for this cell and for this gene. So, for a single-cell analysis of 50 human cells (and there are around 20,000 genes in the human body), we'd have a 50x20000 matrix.
What are Cell Atlases?
So, we now know what scRNA-seq is. What have people done with it? Lots of things, such as analyzing transcriptional changes that occur as we age, understanding cell differentiation in embryos, and even understanding how tumor cells could be better attacked.
But, amongst the most ambitious tasks that scRNA-seq is supporting is in the creation of 'Cell Atlases'. These are terabyte-sized databases, ran by international consortia, with hundreds of institutions and thousands of scientists participating. Contained with them are the transcriptional landscapes of hundreds of thousands of individual cells, selected from an extraordinarily diverse set of tissues (tongue, cerebellum, ovaries, and dozens more), cataloged for public usage. As of this writing, there are comprehensive cell atlases for humans, mice, nematodes, primates, zebrafish, fruit flys, and many more; many of them overlapping in scope, and some of them even mapping fetal or diseased versions of the associated species. Instead of one-off scRNA-seq experiments created for the purposes of single experiments, this was a more concentrated effort, multiple parties divvying up a 'transcriptome map' amongst each other, exploring each one, and combining it all back together.
Coincidentally, the human cell atlases exactly solve the goal we stated at the start of this blogpost: learning the cellular identity of every cell in the human body. Well, we haven't got at ‘every’ cell, but we’ve definitely completed the 80 part of the 80/20 rule and then some. But, while we've pursued our goal purely for knowledge sake, these cell atlases have much loftier visions. The core idea behind them and why they were publicly released is in hopes that scientists around the globe could use them to find better protein targets for tissues of interest, understand how diseased cell states different from healthy ones, predicting medication response, and uncover new proteins/genes/cell types entirely. These atlases are, in many ways, this generations Human Genome Project; an extraordinarily expensive and challenging biological data collection mission, with grand dreams for how it could be used to advance human health in the future.
The problem with scRNA-seq
Have the expectations for these atlases paid off? Lets ask an even more basic question, has scRNA-seq paid off? It's...hard to tell.
On one hand, the citations of Tabula Sapiens, the largest known human cell atlas, certainly does champion the utility of the atlas in helping further biology research. Multiple papers do reference it to confirm suspicions they had or as a secondary source of truth for their own scRNA experiments. And, to be fair, it was only released in 2022, and good biology research does take time.
On the other hand...has it been instrumental to anything? Has there been anything useful we've found purely because of these atlases? I'm absolutely positive there are people who'd define the results we've gotten from scRNA studies as useful, but I'm personally skeptical.
A mildly interesting reddit thread about the whole topic can be found here, where the OP says:
Every day, there is another bloody paper on single cell this, single cell that. All big papers, all big results, all big data, and all came up with the same conclusion: there's a lot of heterogeneity between cells, that cells are different, that at different locations, cells express different genes, that at different stages, cells express different genes, that a clump of extracted tissue contain different cell types and not a homogenous group "like we thought". Like no shit!! And then they dump all of those data in some repository and move on sequencing something else.
While their frustration is more dedicated to scRNA-seq, I'd imagine it applies even more-so to cell atlases; an immense amounts of data, but little follow-up on what exactly any of it means. Others in the thread point out this is somewhat uncharitable, this is simply how science works, but still admit a kernel of truth in the OP's frustration. Generating vast amounts of single-cell RNA data is quickly becoming easy, interrogating it in any capacity is becoming hard. But why is it hard? There are really only two reasons.
Batch integration. Combining datasets from different scRNA-seq datasets is often challenging due to batch effects; as in, each scRNA-seq dataset is subject to its own unique strangeness due to minor differences in how labs go about things, which can create dramatic differences in the numbers you end up with. There are known methods to help solve this, but going this path is much more an art than a science. For example, let's say we collect a dataset of neuron cells from patients with a rare neurological disease, and would like to know the transcriptional changes these neurons have compared to healthy neurons. But the numerical 'space' in which the healthy transcriptomes live will, always, unfortunately be different from those of the diseased neurons. There are ways around this, there's a bevy of batch integration models, but doing that is much more an art than a science. While cell atlases were actually mildly supposed to solve this problem by offering an all-in-one dataset, all subject to the same set of batch effects, realistically the problem still pops up, as it takes months to produce this data + multiple institutions, each with their own processes.
Cell annotation. Typical cell atlases annotate cells based on whether they express certain 'marker genes' (e.g, neuron marker genes), and, for the cells that lack any markers/have multiple markers, they are annotated according to their transcriptomic 'proximity' to these cells with more cleanly defined types. This is related to the data integration problem! Even if you can remove batch efforts from your dataset in order to compare it to the atlas, the basis upon which you compare cells is often based on their cell type! And cell type annotation may sometimes fail for even well-known cell types, and be even more unreliable for rare or entirely unknown cell types.
If you completely solve both of these, scRNA-seq becomes far more useful. Keep in mind, the first order impact of solving these two issues is just speeding up typical analyses, instead of relying on a custom-tuned batch effect corrector algorithm, you simply have a method that 'corrects' everything and you can go on with your analyses as normal. We may even discover some new rare cell types that would go typically unnoticed in usual algorithms.
This is all well and good, but it feels marginal, a mild improvement to a field that hasn't had much impact on human health. But the second order impact is much more exciting: the potential for a computational model of perturbation on cell state. If we have a model that has such a generalized understanding of transcriptomes that it is capable of translating them all to the same numerical space, we can naturally assume it understands much more about a cell than just how to do this simple mapping. It understands how excessive sugar changes cells using what its seen from T2 diabetes patients. It understands how mechanical stress changes cells using what its seen from hypertrophic cardiomyopathy patients. In short, it has learned a sense of how perturbations affect cell state. Such a model could move beyond the datasets it was trained on. It could accurately predict how certain drugs will alter cell states before its ever left a lab, be able to understand the possible evolutionary trajectories of a tumor, or be able to hallucinate whole transcriptomes for rare disease patients that we obviously cannot biopsy in full.
While the cell atlases they themselves haven't been particularly useful, they may end up being useful in a completely unexpected way: as fuel for an engine.
scRNA Foundation Models
History
Foundation models are, in a general sense, 'any model that is trained on broad data (generally using self-supervision at scale) that can be adapted (e.g., fine-tuned) to a wide range of downstream tasks'. They've become most popular in the space of language, but have extended towards different modalities beyond those, including pixels (DALL-E), audio waveforms (MusicGen), and even amino-acid residues of proteins (ESM2). And soon, they began to be applied towards single-cell transcriptomes.
We should first motivate why they were created at all. As mentioned before, the utility of scRNA is massively dragged down by being unable to easily integrate scRNA datasets together and the analyses behind these datasets often relying on fuzzy labels (cell annotations). scRNA foundation models potentially solve both of those in one fell swoop by attempting to map gene count vectors to the same embedding space in a zero shot manner. If we can pull this off, batch effects (mostly) cease to be a problem and we can entirely move beyond cell types annotations as the basis of cell-to-cell comparisons -- if the embeddings represent individual cells well enough, those can be relied upon instead!
It's a simple idea and, as with almost every other concept, it has of course been extensively tried before, with the concept reaching back to the early 2010's; foundation models certainly did not invent the concept of embedding. But it was often limited in its scope, relying on simple linear embedding schemes, being specific to the exact perturbation it was trained on, or simply being trained on a relatively small dataset. A very nice blogpost from markov.bio about scRNA models discusses these shortcomings more in depth. There were a few papers that poked at non-linear data transformations trained on diverse datasets, such as scVI, but the results were still in a hazy territory of being interesting as a paper, but still on shacky ground for being truly useful.
But with the Attention Is All You Need revolution and the increasingly fervorous belief that huge models and huge amounts of data could lead to extraordinary models, biologists started to set up their own single-cell foundation models. As of March 2024, there are quite a few: scFormer, scFoundation, GeneFormer, scBERT, scGPT,, Universal Cell Embeddings (UCE), and several others. Remember GeneFormer, scBERT, scGPT, and UCE, they'll come up later. And the fuel behind how they could be created at all is something we've talked about: cell atlases. Each one of these models rely on multiple of such atlases as training data, using on the immense diversity of cells that have been catalogued and studied for the last decade in order to, hopefully, gleam a finer understanding of cell state than any other model ever could.
How are they? Have the effort we've put into creating these immense cell atlases paid off?
The Problem With scRNA Foundation Models
Of course, everything in biology that people have poured blood, sweat, and tears into have some sort of problem. There are two preprints assessing the current state of scRNA foundation models, both of them funnily enough released within 5 days of each other in October 2023.
The first one is Assessing the limits of zero-shot foundation models
in single-cell biology. They specifically focus on studying scBERT and GeneFormer claims of their 'zero-shot' performance. Zero shot performance here is truly what we care about, if we wanted to dip our toes into having to fine-tune a model for every one of our scRNA problems, the primary point of using a foundation model in the first place is lost! And, unfortunately, they find that while both scBERT and GeneFormer perform decent in zero-shot settings, they are still no better than baseline methods for embedding separation1, batch integration and cell annotation2 tasks. The footnotes share some concerns I have with the metrics, but in all of these, the performance of the foundation models were reasonably similar to the aforementioned scVI. Much more concerningly, simple Highly Variable Genes (HVG's), which are calculated in an entirely non-parametric way, were reasonably competitive with the computationally expensive transformers (Fig 2 and 5) Does this imply that the heavy pretraining that goes into these foundation models are largely useless?
In one of the most unexpected results I've seen in a paper, the answer is yes, at least for scGPT. The proof for this is a little involved: it compares the MSE loss for scGPT versus the MSE loss of predicting the mean expression value for each gene for the masked-language-modeling loss it set up. Astonishingly, the MSE between the two are largely the same across all pre-trained models of scGPT (scGPT random is not pre-trained, so MSE is obviously quite high). The conclusion here is that pre-training is imparting no more useful information than simply being able to parrot the average transcript expression.
So, things are not looking great. Five days later, another paper titled A Deep Dive into Single-Cell RNA Sequencing Foundation Models was released. Here, it looked at two models again: the same scGPT as before, but also GeneFormer. The results from the last paper here are largely recapitulated, but in different flavors. In one task, cell annotation results between zero-shot scGPT mappings and a trained logistic regression model had mixed results:
The authors of scGPT evaluated the model’s cell type annotation capabilities on three different datasets: multiple sclerosis [19], pancreas [20], and myeloid data [21...We found that for the multiple sclerosis data, scGPT outperformed logistic regression; for the myeloid data, logistic regression outperformed scGPT; and for the pancreas data, the two methods performed similarly (Figure 3; Supplementary Table 4).
And, even in few-shot regimes, where scBERT and scGPT were allowed to see a fraction of the data, logistic regression still was competitive or outright outperformed it.
Finally, they also perform a few experiments suggesting that pre-training in scBERT and scGPT is of questionable importance for cell annotation tasks, with it being largely unimportant in scBERT, and of variable importance in scBERT. This is massively distinct from NLP, where strong pre-training performance correlates quite well with downstream task performance. However, unlike the prior paper, they offer some hope that the value of scRNA foundation models is not in solving the typical challenges posed by the scRNA community (such as cell annotation), but in more complex ones. Here's a section from their discussion section which I quite liked:
These results underscore that cell type annotation is not a challenging enough task on which to demonstrate the potential value of single-cell foundation models. The value of foundation models for single-cell RNA sequencing data will instead be showcased through superior performance at more challenging fine-tuning tasks which simple models currently cannot solve, or else through the models’ abilities to capture and elucidate meaningful gene-gene interactions through their learned attention weights. When claiming the latter, it is important to keep in mind that strong downstream performance does not necessarily imply rich representation learning...
I mentioned earlier to remember four model names, but I've only mentioned three so far, scGPT, scBERT, and GeneFormer. What about the fourth?
Universal Cell Embeddings
I'm making a specific section for this paper because I think it'll become a really, really important paper for people to build off on.
Universal Cell Embeddings, or UCE, is the last-to-be-released of all these methods, released as a pre-print in November 2023, whereas the rest of these methods were originally released in mid-2023/2022. There are also a few things unique about this model worth discussing:
It is cross-species. Whereas all of the other discussed models are human-specific, this one includes datasets from 8 species in total in its training data: human, mouse, lemur, zebrafish, pig, rhesus macaque, crab eating macaque and western clawed frog.
It makes clear that it is not meant to ever be fine-tuned. It goes so far as to say 'UCE is able to map any cell, from any 103 tissue[s], or any species, into one shared universal space, with no additional training'. While no foundation model we've discussed thus far has explicitly relied on fine-tuning, this is the only one that proudly eschews the need for it.
It was sponsored by the 800-pound gorilla of cell atlases dataset creators: Chan Zuckerberg BioHub, of Tabula Sapiens, Tabula Muris, Tabula Muris Senis, Tabula Microcebus, and CellXGene fame. If anybody is good at creating scRNA data and open sourcing it, it's them.
How's UCE's performance? Impressive!
It outperforms all the other methods to some degree in typical scRNA analyses tasks, including ones that require fine-tuning:
We compared several methods and found that UCE substantially outperforms the next best method Geneformer by 9.0% on overall score, 10.6% on biological conservation score, and 7.4% on batch correction score (Supplementary Table 1). To comprehensively assess the value of these zero-shot embeddings, we also compare UCE to fine-tuned methods that are conventionally used for this task. Notably, UCE even performs slightly better than non-zero shot methods that require dataset-specific training: scVI and scArches.
But the main interesting part about UCE is how it leverages the unique zero-shot generalization capabilities of foundation models to work on things that other models massively struggle with, not focus on tasks that most models are already quite good at. And it presents three:
The first task is UMAP'ing embeddings of cells collected from an entirely new species unseen by UCE! Due to the UCE being cross-species, this is actually possible! Of course, embedding plots are always going to be a bit controversial, but it's undeniable that there's something going on here; they show reasonable-looking separation of cell types for a primate (suspect because there are other primates in the training dataset) and a chicken (less suspect because there are no other birds in the training dataset). They offer further proof of the validity of their blind cross species annotations here, by the same lead author, released in February 2024.
The second task is attempting to annotate rare cell types. They set up an example workflow where they take an scRNA dataset of a brand new discovered cell type (called Norn cells), do a zero-shot embedding of it through UCE, and train a simple binary classifier to tell between the Norn cell embeddings and not-Norn cells, using the original Norn cell papers dataset for the latter dataset. Then, we simply apply this classifier to our dataset of 36M historically collected cells included in UCE, which is simple because each of the 36M are also made up of UCE embeddings. And viola, we can find Norn cells scattered throughout our old dataset.
The third and final interesting task is interrogating gene expression differences in disease populations (COPD and IPF) versus healthy ones. What's fascinating here is that this analysis was done using the Norn-annotated cells from earlier, which are unconfirmed, but show the expected differential gene expressions between diseases!
(d) Cells predicted to be Norn cells within a lung disease dataset express known Norn markers, as demonstrated by log fold change (LFC). Differential gene expression in predicted Norn cells, grouped by disease status. There are significant differences in gene expression of important Norn markers and genes involved in the production of erythropoietin (Epo) between cells from IPF, COPD and control patients. Patients with IPF and COPD are known to have elevated levels of blood stream Epo, with COPD patients having greater bloodstream Epo levels than patients with IPF.
Can all of these tasks be done without UCE? Of course, and there are plenty of papers that can poke at each of these tasks. But at this ease, scale, and all-in-one? At that point, we're getting into territory that the UCE is flexing the strengths that foundation models are uniquely suited to. Will UCE truly be the winner here? The Alphafold-esque model that reigns strong even after 3 years? I very much doubt it, it may well be the case that scBERT, scGPT, or one of the other models developed in the past are actually the best architecture choice, but UCE is by far the most ambitious model here and should be given some kudos for thinking beyond typical scRNA workflows.
What does the future look like?
Next-generation-sequencing made it possible for the scale of scRNA-seq data to explode. But, as I've mentioned earlier, measuring mRNA transcript levels is a few steps removed from the puzzle of proteins, and even proteins are removed from the more ambiguous question of cell state. As we get better and better at creating these ultra-large datasets, it's easy to imagine a world in which these foundation models no longer operate on RNA alone, but on every proxy of cell state simultaneously: metabolomics, glycomics, proteomics, epigenomics, and, likelier earlier than later, spatial transcriptomics. These sorts of models are popping up all over the place, like this recent paper on a foundation model that uses chromatin accessibility data to learn transcription regions gathered by ATAC-seq and even UCE relies on the famous proteomics ESM2 paper for some of its pre-training, and its only a matter of time before these are all slowly merged together to take advantage of every possible token.
The future will be very interesting!
This is a strange metric in my opinion. The paper says, ‘One key aspect of evaluating cell embeddings is the degree to which cell types are distinct within the embedding space.’ This implies that you can actually trust cell types, or that some notion of fuzziness in cell types doesn’t exist! Embedding utility is of course important, but why not use perturbation datasets for this?
The way they measured this is extremely qualitative and…hard to know if it means anything. It’s literally just eyeballing: ‘As commonly done in single-cell transcriptomics, we used UMAP projections to visually inspect embeddings (Fig. 4). By annotating the UMAP by cell type (Fig. 4A) versus experimental technique (Fig. 4B), we jointly assess if cell embeddings correct for batch effects stemming from techniques while still retaining cell type identity…..Overall, we observed that while Geneformer and scGPT-human can integrate different experiments conducted with the same experimental technique, they generally fail to correct for batch effects between techniques. As depicted in Fig. 4A, the cell embedding space generated by Geneformer fails to retain information about cell type, and any clustering is primarily driven by batch effects (Fig. 4B). On the other hand, the space created by scGPT offers some separation of cell types (Fig. 4A), but the primary structure in the dimensionality reduction is driven by batch effects (Fig. 4B). In contrast, even the simple baseline of selecting highly variable genes (HVG) qualitatively produces a similar or better result to scGPT, with the Smarter technique now being integrated with InDrop. Finally, we observed that scVI mostly integrates this dataset, forming clusters primarily due to cell type, with most techniques in the same cluster.’







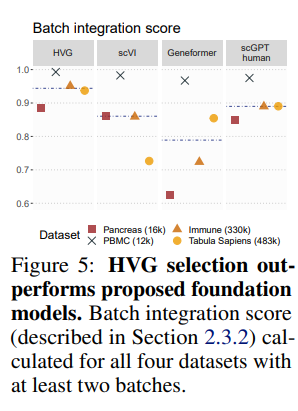







It’s interesting that the cross-species UCE model is the most performant. InstaDeep recently published their ChatNT genomic model and showed that training on multiple species genomes simultaneously, and selectively switching between objectives, produced a model that generally outperforms specialized models. Makes me wonder whether providing several “escape hatches” in terms of objectives makes the training process more robust to getting stuck in local minima.
Hello Abhishaike,
I really enjoyed reading this post. I'm particularly excited about it because I'm preparing for an upcoming STEM outreach program for high school students on the applications of AI in single-cell biology. With your permission, I'd love to incorporate some of the points you discussed in my presentation.